


Research
SCROLL DOWN
Research
Overview
The Khavari lab studies genome regulation in stem cell differentiation and cancer. The basis for how stem cells decide to either differentiate or become cancer is our major focus. We use genomics and proteomics-based approaches combined with versatile genetic models and biocomputation to identify new regulators and networks that regulate these processes. Integral to these efforts is development of new molecular therapeutics for human disease.

Genome Regulation
Genome Regulation
Signaling, LncRNAs, and Genome Regulation
Signaling pathways integrate extracellular signals to productively reprogram genomic expression. With respect to signaling, the lab focuses on essential pathways in epidermal stem cell differentiation and cancer. These include the Ras-Raf-Mek-Erk MAPK pathway and its interactions with other specific pathways, including those involving PI3K, Notch, and WNT. Work in the lab has been the first to knockout the Ras-Raf-Mek-Erk MAPK pathway in mammals [Developmental Cell (2007)] to demonstrate its essential role in tissue homeostasis, the first to define the genomic programs induced in early cancer progression triggered by oncogenic Ras activation [Cancer Cell (2009)]as well as the first to characterize the dependence of the pathway on the IQGAP1 scaffold in cancer [Nature Medicine (2013)].
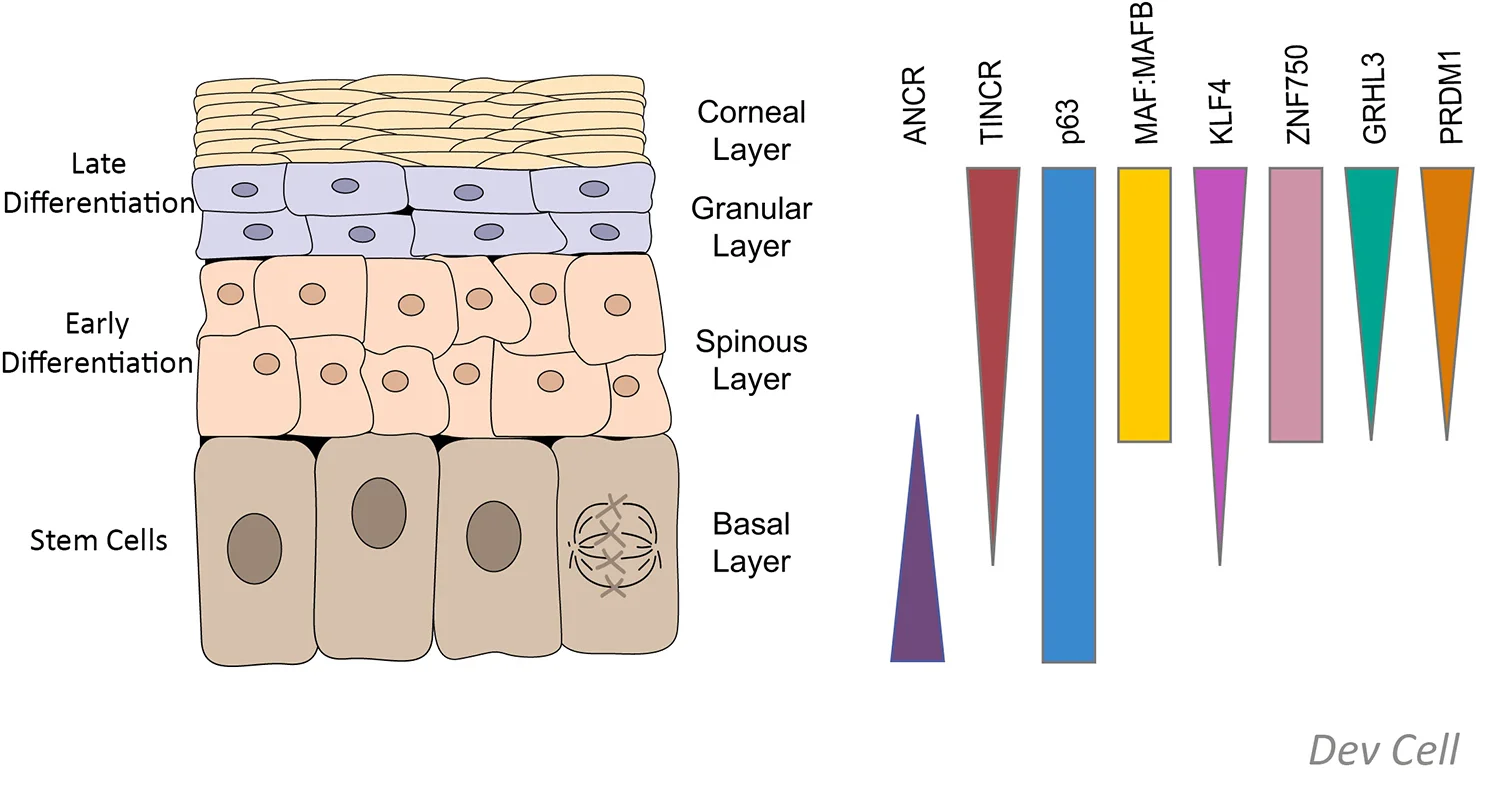
In the case of long-noncoding RNAs (lncRNAs), the lab discovered the first lncRNAs involved in somatic stem cell differentiation, including the ANCR stem cell lncRNA [Genes & Development (2012)] and the terminal differentiation lncRNA, TINCR [Nature (2013)]. Discovery and integration of new roles for signaling networks, epigenomic modifiers, TFs, and lncRNAs is a major focus of the lab designed to define essential regulators of stem cell differentiation and cancer.
For genome regulation, the lab focuses on how essential TFs and lncRNAs interact with regulators of DNA methylation and chromatin to control genome expression in stem cell differentiation and cancer. Work in the lab was the first to demonstrate an essential role for DNA methyltransferases [Nature (2010)] and for the EZH2-JMJD3 histone methyltransferase-demethylases [Genes & Development (2008)] in tissue homeostasis. The lab has also identified TFs with new roles in stem cell differentiation, including ZNF750 [Developmental Cell (2012), Genes & Development (2014)] and MAF/MAFB (Developmental Cell (2015)].

.
.
Innovation Focus Areas
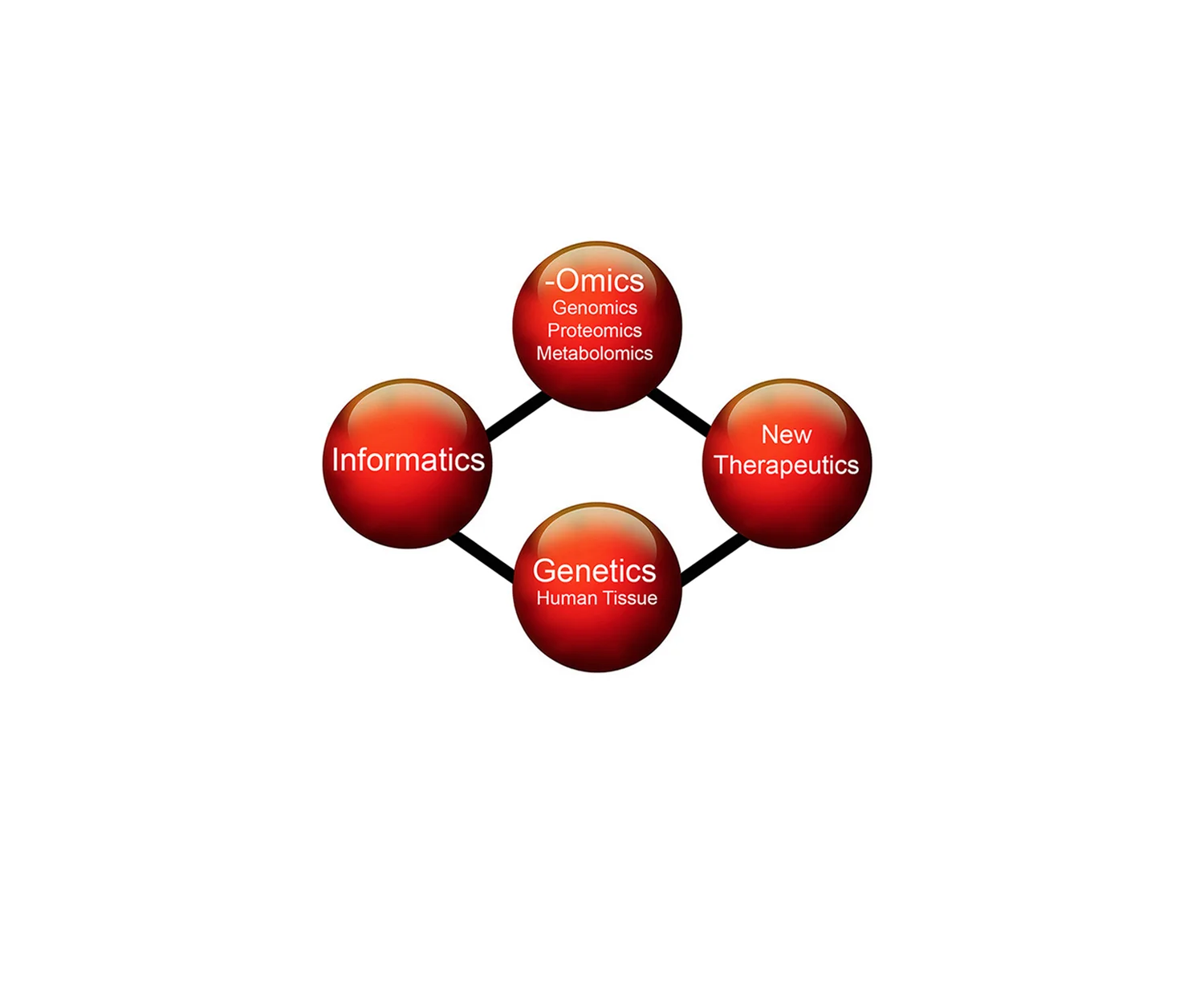
The lab’s innovation focus areas include Genetics, -OMICs, Informatics, and New Therapeutics.
Lab innovation in GENETICS focuses on developing new approaches in 3-dimensionally intact normal human tissue in which multiple genes have been altered. Recent lab innovations have altered 4 or more alleles using multiplex and CRISPR-based approaches in veridical models of stem cell differentiation [Nature (2013), Cell Stem Cell (2013), Developmental Cell (2015)] and cancer [Nature Medicine (2010), Nature Medicine (2013), Nature Genetics (2014)]. These powerful new human tissue models are designed to apply the precision and versatility of functionator organism models, such as yeast and flies, with the complexity and clinical relevance of human tissue.
Lab innovation in –OMICs focuses on developing new methodologies to elucidate the biological networks that govern stem cell differentiation and cancer. These methodologies are designed to shed new light on both the mechanisms of genome regulation as well as post-transcriptional control mechanisms. Recent lab innovations generated RNA interactome analysis (RIA) to define the physical network of RNAs that interact with a given RNA of interest [Nature (2013)], RNA-protein microarray analsysis (RP-PMA) to define the spectrum of protein interactions for a given RNA [Nature (2013), BMC Genomics (2013)], and on-plate ATAC-seq to enable efficient genome-wide profiling of open chromatin in adherent cells [Cell Stem Cell (2013)].
Lab innovation in INFORMATICS has been a central effort to both analyze new data types emerging from in-lab –OMICs innovation as well as datasets generated outside the lab by conventional approaches, including data from the ENCODE project. These methodologies are designed to facilitate high dimensional data analyses that provide an integrative picture of the biologic networks that control tissue homeostasis and cancer. Recent lab innovations include RINNS to efficiently identify non-human sequences in RNA-seq data [Bioinformatics (2012)], FOCIS to integrate ChIP-seq information for identification of co-binding TF enrichment scores [Genome Research (2014)], new ranking analysis algorithms for RIA-seq and RP-PMA data [Nature (2013)], and multiple variant caller pipeline integration for genomic sequencing in human cancer [Nature Genetics (2014), Nature Genetics (2015)].
Lab innovation in THERAPEUTICS focuses on both inherited human epithelial diseases caused by single gene mutations, such as epidermolysis bullosa (EB), as well as cancer. Innovations have included epidermal stem cell targeting for corrective gene transfer that has led to a phase I clinical trial in humans [Human Gene Therapy (2010)] as well as identification of new approaches to disrupt signal transmission in cancer by altering interactions between the Erk1/2 MAPK pathway and relevant scaffold proteins [Nature Medicine (2013)]. Therapeutics efforts leverage insights into stem cell differentiation and cancer with innovations in the other 3 innovation focus areas to develop new molecular treatments for human disease.
